Single-cell mrna transfection studies: delivery, kinetics and statistics by numbers
Single-cell mRNA transfection studies: Delivery, kinetics and statistics by numbers Carolin Leonhardt a, 1, Gerlinde Schwake a,1, Tobias R. Stögbauer, PhDa, Susanne Rappl a, Jan-Timm Kuhr, PhDb, Thomas S. Ligon, PhDa, Joachim O. Rädler, PhDa,⁎ aFaculty of Physics and Center for NanoScience (CeNS), Ludwig-Maximilians-Universität, München, Germany bInstitut für theoretische Physik, Technische Universität Berlin, Berlin-Charlottenburg, Germany Received 15 March 2013; accepted 18 November 2013 In artificial gene delivery, messenger RNA (mRNA) is an attractive alternative to plasmid DNA (pDNA) since it does not require transfer into the cell nucleus. Here we show that, unlike for pDNA transfection, the delivery statistics and dynamics of mRNA-mediated expression are generic andpredictable in terms of mathematical modeling. We measured the single-cell expression time-courses and levels of enhanced green fluorescent protein(eGFP) using time-lapse microscopy and flow cytometry (FC). The single-cell analysis provides direct access to the distribution of onset times, lifetimes and expression rates of mRNA and eGFP. We introduce a two-step stochastic delivery model that reproduces the number distribution ofsuccessfully delivered and translated mRNA molecules and thereby the dose–response relation. Our results establish a statistical framework formRNA transfection and as such should advance the development of RNA carriers and small interfering/micro RNA-based drugs.2013 The Authors. Published by Elsevier Inc. All rights reserved.
Key words: mRNA transfection; Non-viral gene delivery; Expression kinetics; Single-cell studies; Pharmacokinetics in gene therapy applications. Firstly, mRNA does not requiretransfer into the nucleus and hence mRNA transfection is also Nucleic acid transfer is widely used in basic research as well as effective in non-dividing cells, which is a major drawback of biomedical applications. In recent years, novel stabilized mRNA pDNA transfectionThis makes mRNA a particularly strong constructs have become more prevalent in therapeutic applications therapeutic agent in dendritic cells which are otherwise hard to showing superior properties compared to plasmid DNThis transfectSecondly, immunogenic response to mRNA progress is mostly due to the discovery of 5′ mRNA anti-reverse activated by Toll-like receptors (specifically TLR3) is less cap analogues (ARCA), to the insertion of additional untranslated pronounced compared to unmethylated CpG motifs of DNA regions, and to poly(A) tails that significantly promote and prolong recognized by TLR9In addition, mRNA transfection efficient translation of foreign mRNA inside cellsIn general, remains transient, preventing the risk of permanently integrating mRNA delivery has considerable advantages over pDNA delivery into the genome. Hence, mRNA delivery is of increasing interestfor future biomedical applications in particular with regards tostrategies that aim to use mRNA as a programmable device for This is an open-access article distributed under the terms of the Creative controlled intracellular mRNA targeting and in situ logic Commons Attribution-NonCommercial-ShareAlike License, which permitsnon-commercial use, distribution, and reproduction in any medium, provided evaluation of disease-related conditions the original author and source are credited.
The major hurdle to clinical trials remains the delivery of Financial support by the Elite Network of Bavaria is gratefully nucleic acid to eukaryotic cells. As a result, an ongoing search is acknowledged by CL. This project was supported by the German Excellence still underway for non-viral delivery methods that are optimized Initiative of the Deutsche Forschungsgemeinschaft (DFG) via the Excellence for efficient and controlled delivery of mRNA. Since the first Cluster "Nanosystems Initiative Munich" (NIM), the Sonderforschungsbereich non-viral delivery of mRNA using cationic lipids by Malone, "Nanoagents" SFB 1032, and by the EU-FP7 project ‘‘NanoTransKinetics".
Felgner and many synthetic delivery systems were E-mail address: (J.O. Rädler).
found to be effective for mRNA delivery, with generally better 1 These authors contributed equally to the work.
efficiency found for liposomes than for It is 1549-9634/$ – see front matter 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Please cite this article as: Leonhardt C., et al., Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine: NBM2014;xx:1-10,
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
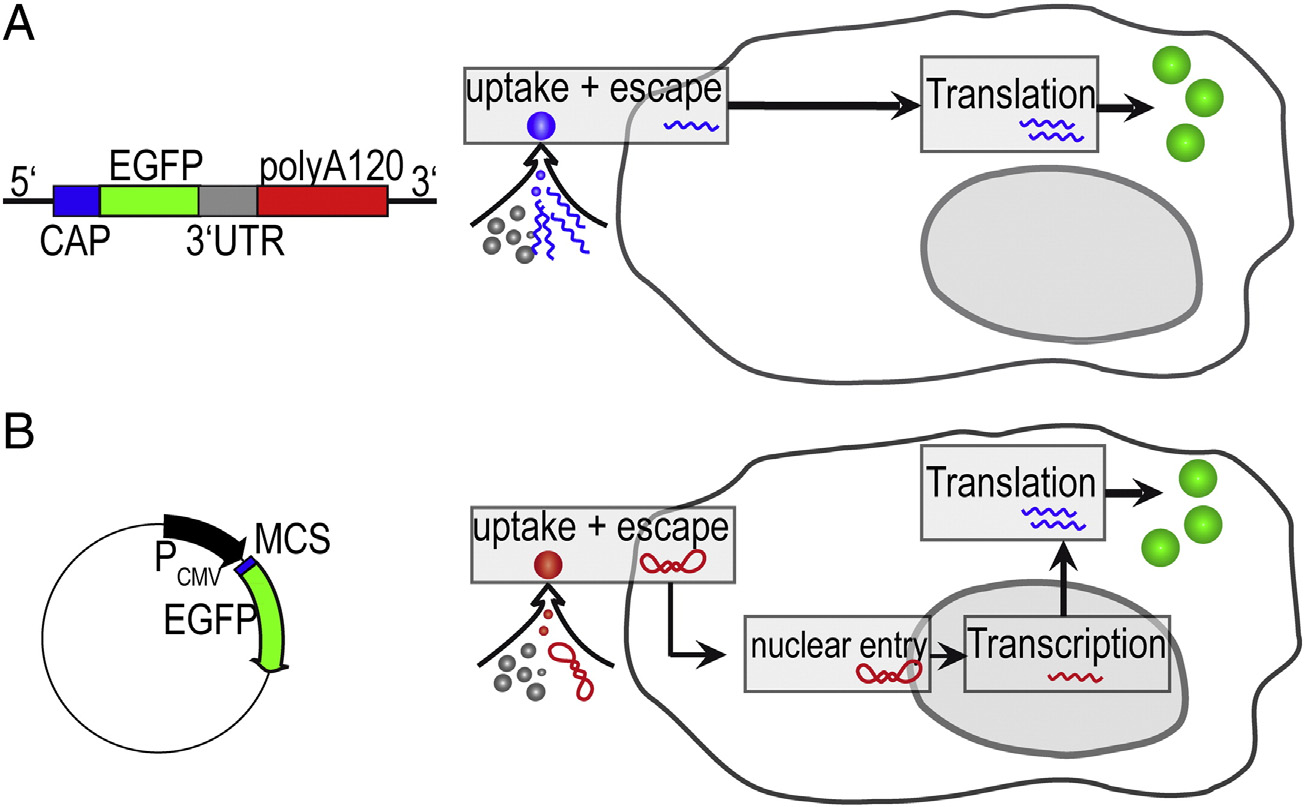
Figure 1. Comparison of mRNA and pDNA Vectors (both gene vectors encoding for the same eGFP protein) and their respective uptake pathways. (A)Linearized RNA (1192 bases) furnished with a stabilizing CAP sequence, an enhancing UTR sequence, and poly-(A) tail. (B) pDNA (4733 base pairs) under thecontrol of the CMV promoter. The vector transfer under identical transfection protocols differs because mRNA is translated after endosomal escape, whileplasmid DNA must be transferred into the nucleus for the initiation of transcription.
generally accepted that both mRNA as well as pDNA are
mRNA vector encoding for eGFP. Single-cell fluorescence time-
translocated via endosomal uptake, cytosolic release and - in case
courses were fitted based on rate equations for translation and
of pDNA - nuclear entry. However, mechanistic insights are
mRNA/eGFP degradation yielding the onset time distribution,
mostly limited to assessment of changes in the transfection
mRNA/eGFP degradation rates, and the expression rate. The
efficiency as a function of biochemical or structural variations of
mRNA expression model applies to at least three different cell
the carrier. A full pharmacokinetic model, which in principle has
lines. We interpret the cell-to-cell variability in eGFP levels, i.e.
been established using compartment models and rate
the distribution of expression rates, in terms of number of
lacks validation due to the multitude of kinetic
successfully delivered and translated mRNA. The latter is
rates. In comparative studies, it was shown that mRNA
estimated using a two-step stochastic delivery model. The model
transfection compared to pDNA transfection is faster and yields
assumes delivery of mRNA in finite size complexes that are
a larger fraction of transfected However, a more
taken up stochastically by endosomes and randomly released
detailed and quantitative understanding in particular of artificial
from endosomes into the cytosol. The model quantitatively
mRNA delivery is of increasing importance for gaining a
reproduces the dose–response relation and yields the correct
systems-level description of the kinetics of RNA-based
shape of the distribution function. As such, this work represents
deviceThe degree of predictive power describing
an advance in predictive modeling of mRNA transfection for
synthetic RNA expression level and timing will nevertheless
quantitative gene expression studies, which we believe will be
depend on the degree of accuracy with which the transfer
particularly useful for research on siRNA and miRNA kinetics.
efficiency and transfer kinetics can be described. Moreover,predictive modeling of mRNA transfection will be instrumentalfor the advancement of mRNA based therapies. Yet, any non-
viral delivery is inherently stochastic and the expression level
pDNA and mRNA-vectors
and timing of every single cell is different. Hence, measurementsat the single-cell level and analysis of the corresponding
Two different vectors for pDNA and mRNA transfection
distribution functions are necessary to acquire the true
were designed. The peGFP-N1-Vector (commercially available
population response in transfection experiments. Using single-
at BD Biosciences Clontech, Germany, 4733 base pairs) is the
cell analysis, we recently showed that in the case of pDNA
standard eGFP vector. As an mRNA reference construct for in
transfection, the distribution of gene expression levels can be
vitro transcription, we designed a vector that is based on the
reproduced using a stochastic Similarly, a recent
pSTI-A120-vector (4746 base pairs, transcript 1192 bases),
statistical analysis of nanoparticle dosing exhibited Poisson-type
which has previously been described in literaturThe
distribution in the number of nanoparticles being taken
complete vector map is presented in Figure S1. Both vectors
Here, we study gene expression after non-viral delivery of
contain the same eGFP gene but differ in their promoter region:
synthetic mRNA analyzing single-cell expression traces in terms
The peGFP-N1-Vector has a strong CMV-promoter for
of numbers of complexes delivered and numbers of proteins
expression in vitro. The mRNA is generated with a commercial
being expressed. Using single-cell fluorescence time-lapse
in vitro transcription kit from the pSTI-A120-vector under the
imaging and FC, we monitored expression of a cap-stabilized
control of the T7 promoter. The backbone of both vectors is
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
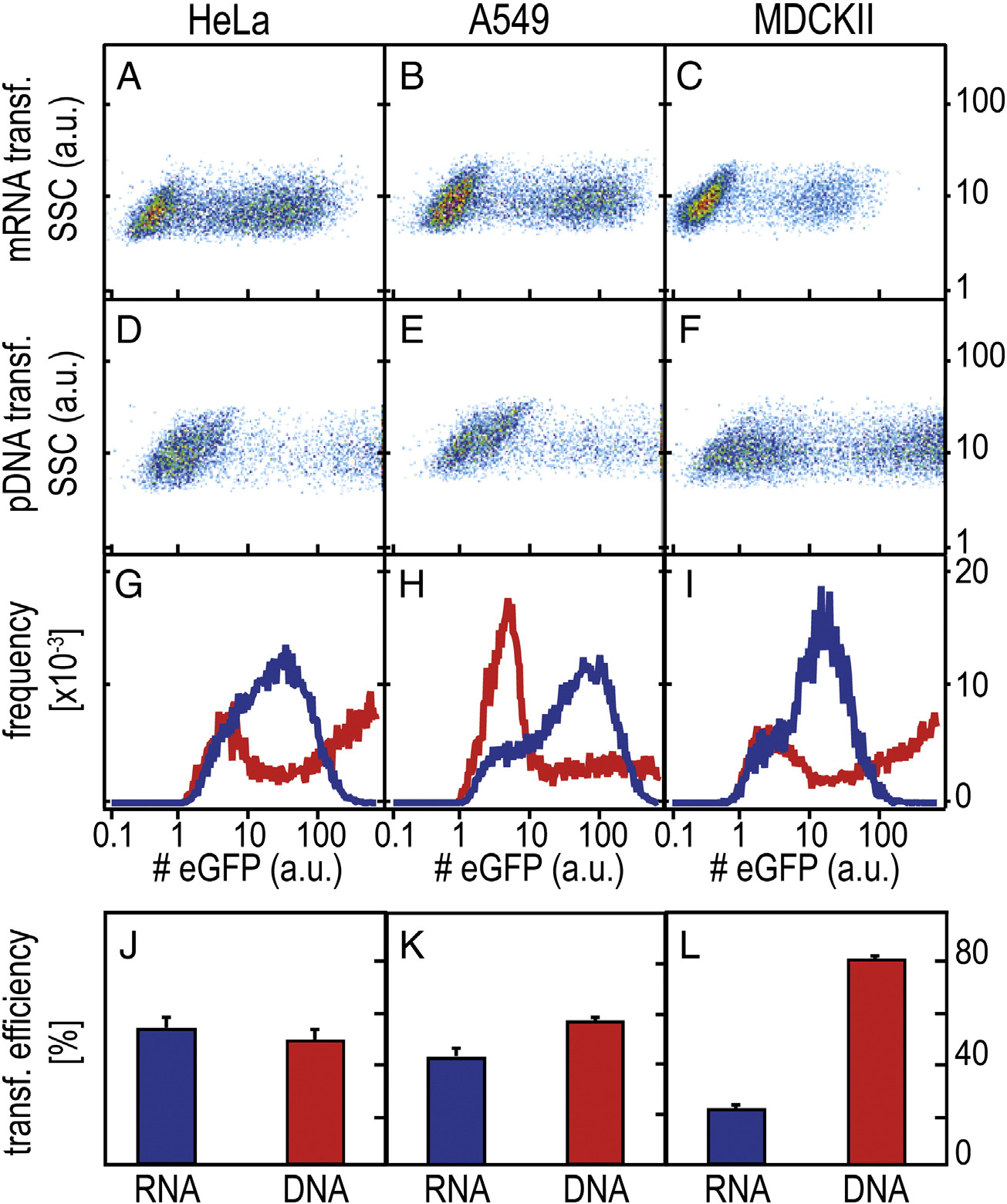
Figure 2. Representative FC scatter plots for mRNA- and pDNA- mediated eGFP expression in three different cell lines (arbitrary units). (A-F) Two-dimensionalscatterplots (sideward scatter vs. fluorescence intensity) for HeLa, A549 and MDCKII cells 25 h post-transfection with mRNA and pDNA. (G-I) Averagefluorescence intensity per fluorescent cell (RNA data are shown in blue, DNA data are shown in red); (J-L) Percentage of fluorescent cells (mean ± SD).
based on the pCMV-Script vector. pSTI-A120 has a 120-bp
from Invitrogen, Germany. Syto RNAselect was purchased from
poly(A) tail and a 3′ untranslated region (UTR) from human β-
Life Technologies, Germany. 6-well culture plates (Falcon) were
globin enabling in vitro transcription of polyadenylated RNA.
purchased from VWR International GmbH, Germany. Sterile
To generate in vitro-transcribed mRNA (IVT RNA), the
PBS was prepared in-house. Ham's F-12K, MEM, DMEM and
plasmid is linearized downstream of the poly(A) tract by SapI
Trypsin-EDTA were purchased from c.c.pro GmbH, Germany.
digestion and purified by phenol/chloroform extraction and
sodium acetate precipitation. One μg of the linearized vector isused as a template for the in vitro transcription reaction using the
A human alveolar adenocarcinoma cell line (A549, ATCC CCL-
Biozym Kit (MessageMAX™ T7 ARCA-Capped Message
185) was grown in Ham's F12K medium supplemented with 10%
Transcription Kit). Having an Anti-Reverse Cap Analog
FBS. HeLa cells (ATCC CCL-2) were cultured using minimum
(ARCA) (m 7, 3′-O
G[5′]ppp[5′]G) cap on the 5′ end, ARCA
essential medium (MEM) with Earle's salts and L-Glutamine
cannot be incorporated in the reverse orientation. Thus, 100% of
supplemented with 10% fetal bovine serum (FBS). A Madin-Darby
the caps in the produced IVT RNA are in the correct orientation,
Canine Kidney epithelial cell line (MDCKII, ATCC CCL-34) was
increasing the translation efficiency of the IVT
cultured in DMEM with 4,5 g/L glucose and 110 mg/L pyruvate,supplemented with 10% fetal bovine serum. All cell lines weregrown in a humidified atmosphere at 5% CO2 level.
FBS, Leibovitz's L-15 Medium (Gibco), Lipofecta-
The cells were transfected with equimolar amounts of pDNA
mine™2000, OptiMEM (Gibco) and Sybr Gold were purchased
and mRNA for FC measurements and with equal weight amounts
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
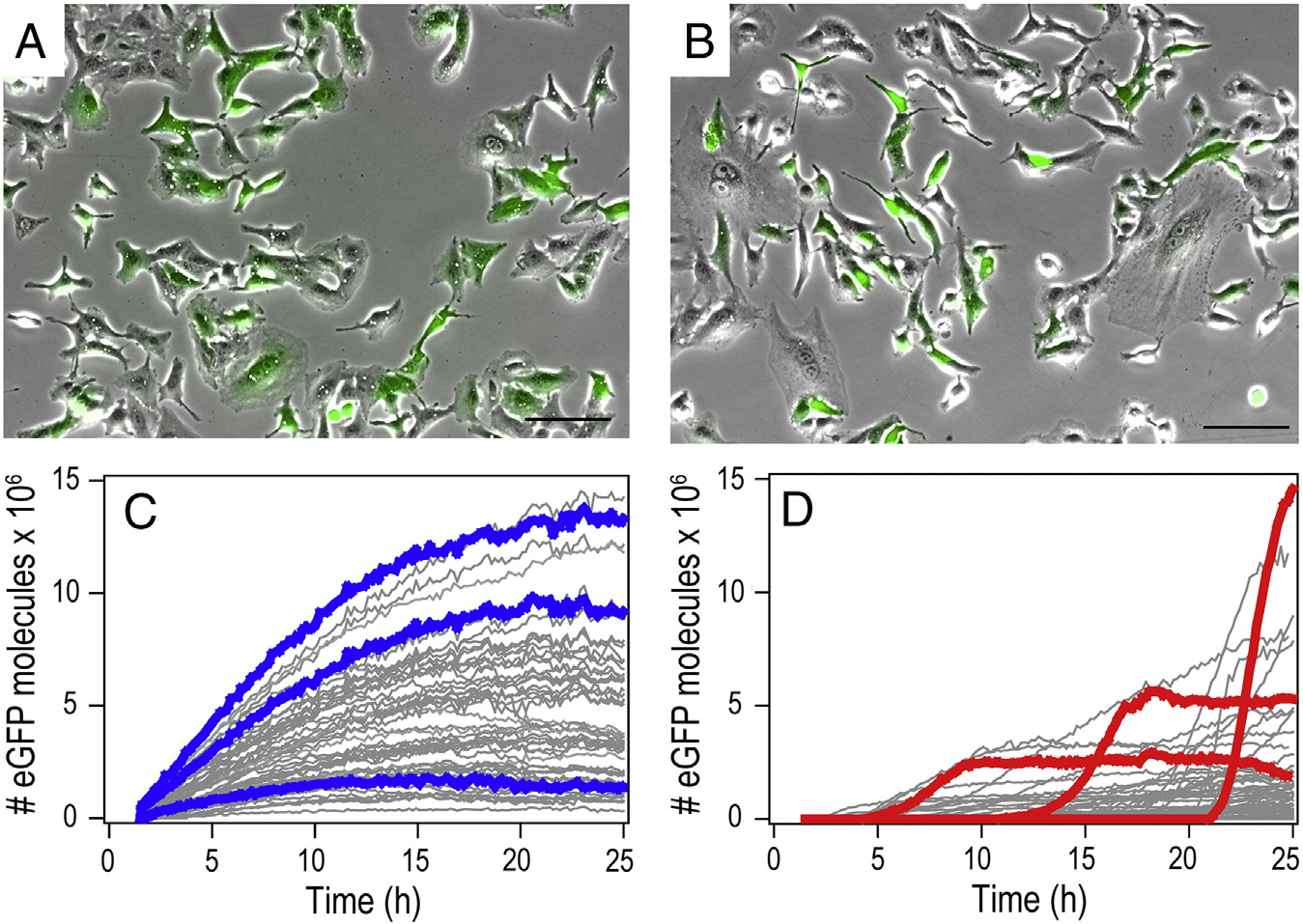
Figure 3. mRNA- and pDNA-mediated gene expression kinetics. (A, B) Exemplary images of an average transfection of A549 cells 25 h post-transfection(overlay of bright field and eGFP fluorescence image. Scale bars 100 μm). (C, D) Representative fluorescence time-courses of eGFP gene expression aftertransfection with mRNA (C) and pDNA (D). To highlight the characteristic differences, we chose and color-labeled three exemplary time-courses each. mRNAexpression shows early onset and continuous rise in the eGFP level, while pDNA expression exhibits delayed onsets and S-shape expression time-courses.
of pDNA and mRNA for single-cell measurements (see
eGFP quantification and calibration
Supplementary). The same transfection reagent (Lipofecta-mine2000®) and the same standard transfection protocols were
To calculate numbers of eGFP molecules from grey values of
used for pDNA and mRNA delivery. For transfection with
the recorded time-lapse movies, a calibration-channel system
fluorescently labelled mRNA, we followed the standard pro-
was developed. Micro channels of known dimensions were filled
tocols for labelling mRNA with Sybr Gold/Syto RNAselect and
with eGFP solutions of defined concentrations. Images of the
prepared lipoplexes with labelled mRNA.
channels were taken under the same experimental conditions asthe monitored expression kinetics data, corrected for backgroundand analysed to get calibration curves. For a detailed description
Data acquisition and quantitative image analysis
of the calibration method, see Supplementary.
Live-cell imaging was performed on a motorized inverted
microscope (Nikon, Eclipse Ti-E) equipped with an objectivelens (CFI PlanFluor DL-10 ×, Phase1, N.A. 0.30; Nikon) and
eGFP fluorescence intensity in cells was measured by FC
with a temperature-controlled mounting frame for the micro-
(Partec, CyFlow space). Flow cytometer settings were adjusted
scope stage. To acquire cell images, we used a cooled CCD
to discriminate transfected and non-transfected cells. The
camera (CLARA-E, Andor). A mercury light source (C-HGFIE
Windows™ FloMax® software package was used for data
Intensilight, Nikon) was used for illumination and a filter cube
analysis. See Supplementary for additional information.
with the filter set 41024 (Chroma Technology Corp., BP450-490, FT510, LP510-565) was used for eGFP detection. Anillumination shutter control was used to prevent bleaching.
Images were taken at 10 fold magnification with a constant
mRNA vs. pDNA transfection
exposure time of 1300 ms at 10-minute intervals for at least25 hours post-transfection. Fluorescence images were consoli-
In a first set of experiments, mRNA-mediated transfection
dated into single-image sequence files. Negative control images
was quantified using FC and compared to pDNA-mediated
were taken to assess lamp threshold values and were subtracted
transfection as a reference. As schematically depicted in
from corresponding image sequence files to eliminate auto-
the design of the mRNA vector A) was chosen for
fluorescence effects. Using SINGLECELLTRACKER, an in-house-
maximal analogy to the pDNA vector. The pDNA vector is a
development software based on fluorescence intensi-
commercial eGFP plasmid equipped with a CMV promoter
ties were integrated over cell contours and corrected for
B). The mRNA construct consists of polyadenylated
background noise. The software calculates the cells' fluores-
RNAs enabling in vitro transcription under the control of the T7-
cence over the entire sequence and connects corresponding
promoter and contains 2 sequential human β-globin 3′UTRs as
intensities to time-courses of the fluorescence per cell.
well as the anti-reverse cap analog (ARCA) (see also
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
the x-axis and the sideward scattering signal on the y-axis showconsistent bimodal populations. Both mRNA and pDNAmediated transfection exhibit eGFP-expressing cells and cellsthat do not express any eGFP. However, for three different celltypes, the fluorescence level of eGFP expressing cells in case ofpDNA mediated expression is more broadly distributed andshifted towards higher values than the eGFP distributionappearing in mRNA transfection. This effect is also seen in theintegrated representation, where the distribution of the averagenumber of eGFP molecules per eGFP expressing cell is shown(, G–I). Here, pDNA transfection is shown in red andmRNA transfection in blue. Note that for pDNA transfection,22% (HeLa), 7% (A549), and 28% (MDCKII) of the cells exhibiteGFP expression levels of 1000 (a.u.) and higher that are notshown for better clarity. In the last row (, J-L), thepercentage of transfected cells are depicted, which is a directmeasure of the transfection efficiencies. We find slightly lowerpercentages of transfected cells for mRNA-transfected cellscompared to pDNA-transfected cells except for MDCKII cells,which feature higher transfection for pDNA vectors.
Single-cell mRNA expression kinetics
The most revealing difference between transfection with
mRNA and pDNA is seen in the single-cell expression kineticsretrieved from time-lapse studies (). Typically, begin-ning after 1.5 hours of incubation, fluorescence microscopymovies were taken over 25 hours using automated time-lapsemicroscopy. The total fluorescence intensity of each single cellwas followed by image and converted into the numberof eGFP molecules per cell (see Supplementary). showstwo typical microscopy images of transfected cells 25 hourspost-transfection (A and B). Bright field andfluorescence images were overlaid to illustrate the fraction oftransfected cells. C and D show gene expression time-courses of single cells. To highlight the characteristic differencesin the expression kinetics, we picked three representative traceseach and show them in color. While mRNA-transfected cellsshow an early and steady rise to a maximum with a subsequentdecrease, pDNA transfection results in sigmoidal intensity time-courses with a steady-state level of eGFP expression and random
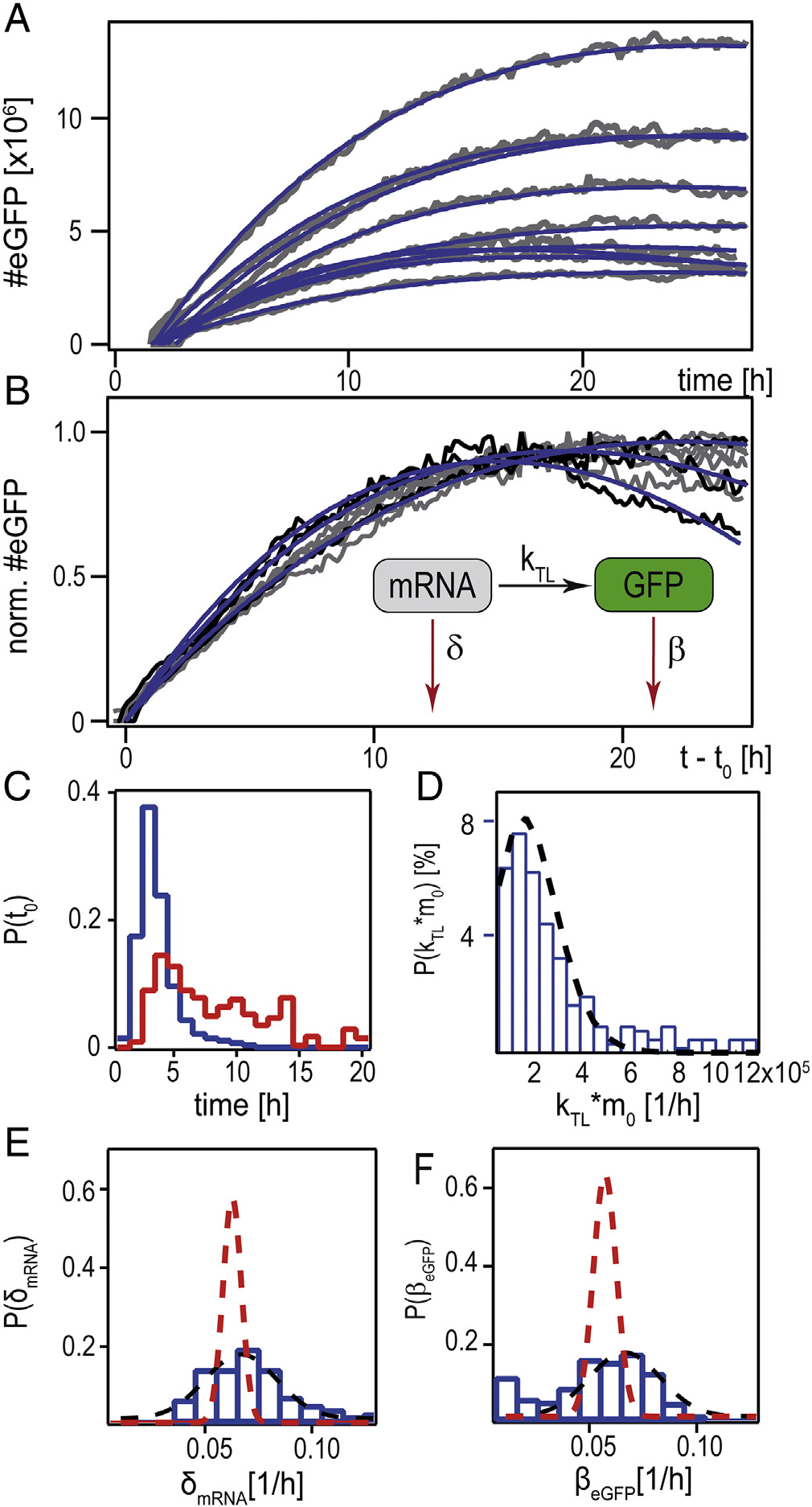
Figure 4. Single-cell mRNA translation, analyzed by a kinetic rate model. (A)
onset times. In contrast to the ubiquitous early onset of eGFP
Time-courses of eGFP expression after mRNA transfection (gray lines). Blue
expression with mRNA that mainly occurs within 5 hours after
lines are fits according to the rate equation model (shown schematically as
transfection, the onset of eGFP expression after transfection with
insert in (B)). (B) Shows the same data as (A), normalized to their maximalvalue and shifted by their fitted onset times, t
pDNA is spread over the range of 2 hours to 20 hours.
0. (C) Distribution of the onset
time t0 (mRNA data shown in blue, pDNA data shown in red). (D)
Modeling mRNA expression
Distribution of the expression rate kTL · m0. (E) Distribution of the mRNAdegradation rate. The black dashed line shows the Gaussian fit to the
Since mRNA transgene expression solely involves transla-
experimental data, whereas the red dashed line is the Gaussian fit to simulated
tion, quantitative modeling reduces to a simple biochemical
data (see Supplementary) (F) Distribution of the eGFP decay rate. Dotted linesrepresent the Gaussian fit to experimental (black) and simulated (red) data.
reaction scheme defined by three kinetic rates as shown in, B. The schematic shows a rate equation model formRNA expression consisting of translation, mRNA, and eGFP-degradation. The model is described by the following set of
To collate the outcome of the transfection
equations for the changes in the number of eGFP molecules,
experiments, identical transfection protocols were followed for
G(t), and the number of mRNA molecules, m(t):
mRNA and pDNA transfection using the commercial cationiclipid agent Lipofectamine2000®.
The FC data shown in were taken 25 hours post-
transfection. The scatterplots with the fluorescence intensity on

C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
course showing an exponential increase with rate δ-β and a long-term decay with decay rate β (see Supplementary). Each fityields an individual set of parameters. C-F presents thecorresponding distribution of the best-fit parameters, which willbe discussed in the following.
Expression onset time distribution
In C, the onset time of mRNA (blue) is shown in
comparison to the onset time for pDNA transfection (seeSupplementary). The faster transfer of mRNA is clearlydocumented in this distribution. In the case of A549 cellsshown here, the onset time distribution after transfection withmRNA peaks approximately 3 hours after transfection andhardly shows any delayed expression onset events after 5 hours,whereas the pDNA onset time distribution is spread over theinterval between 2 and 20 hours post-transfection. The time-distribution is an indirect, yet quantitative measure for thetransfer time of delivery. As known from microscopy studies,endosomal uptake already starts 10–30 minutes afterTherefore, the measured delay in case ofmRNA transfer must be limited by endosomal escape rates.
Remarkably, mRNA expression onset ceases after 10 hours,indicating that no more endosomes lyse or (more likely) thatmRNA molecules are degraded in acidic late endosomes. Thebroadly distributed onset times for pDNA are associated withrare nuclear entry events, which are believed to occurpredominately during mitosis.
mRNA degradation rates
, E shows the distribution of the mRNA degradation
rate retrieved from fitting single-cell time-courses with the
Figure 5. Dose–response relation. (A) Percentage of positively transfected
described model. The average mRNA degradation rate of 0.062/h
A549 cells as a function of increasing amount of mRNA (0.05/0.1/0.5/1/
(corresponding to an mRNA life time of t
2 μg). Squares correspond to FC data. The dashed grey line is a single-
rough agreement with the literature value of The value
Poisson fit, the black line is a double-Poisson fit according to our stochastic
is clearly smaller than the degradation rate of endogenous mRNA
delivery model. (B) Corresponding fluorescence intensity distributions as
measured by FC (bottom to top with increasing mRNA dose).
δ b which is consistent with the reportedly higher
stability of ARCA capped mRNA vectors. The distribution ofmRNA degradation is well described by a Gaussian with half-
width 0.024/h. This variability in the degradation rate is on the
order of the so-called "extrinsic noise" in Thevalues for the degradation of eGFP (with a mean of 0.056/h) are
higher than values that have been reported In
TL denotes the translation rate and δ and β the degradation
rates of mRNA and eGFP, respectively. With t
general, it is noteworthy that the single-cell analysis yields
0 being the time of
expression onset and the initial conditions G (t
estimates for δ and β with high accuracy. The Gaussian fit yields
mean values with less than 6% relative error. Knowing the
the following solution for the number of eGFP
molecules is obtained:
degradation rates is of great value for the improvement of novel
vectors and capping sequences. Furthermore, the degradationtimes are a key to predicting the time-course of expression. In
mRNAðtÞ ¼ kTL⋅m0 ⋅
fact, analysis of Eq. predicts that the maximum of expressionis reached approximately at tmax = 17 h. The time point of half
Applying Eq. to the experimental time-courses, the data are
maximum expression value in the declining late phase of
indeed well fitted. The blue curves in , A show
expression is t1/2 = 45 h. The latter is important because it is a
exemplary best fits to single-cell time-courses (from a total of
measure for the duration of the transient mRNA expression. Note
281 time-courses). There are four free parameters: the onset time
that Eq. also holds for the case δ b β (see Supplementary).
t0, the product of translation (kTL) and initial number of
Moreover, the expression rate kTL · m0 and the difference in
effectively translated mRNA molecules (m0), as well as mRNA
the degradation rates (δ-β) both determining the amplitude and
and protein degradation rates (δ and β). Eq. entails a time-
hence the maximal expression levels, are uncorrelated (see
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
Supplementary, Figure S3C). In E and F, Gaussian fitsto simulated data are additionally shown. For simulation, we usedthe experimentally measured mean degradation rates (seeSupplementary). These fits should represent intrinsic noiseonly, which accounts for about 30% of the total noise. Theadditional width of the experimental data can be attributed toextrinsic sources of noise involved in the gene transfer process.
The kinetics of mRNA proves to be generic because different celltypes show the same mRNA expression curves (seeSupplementary).
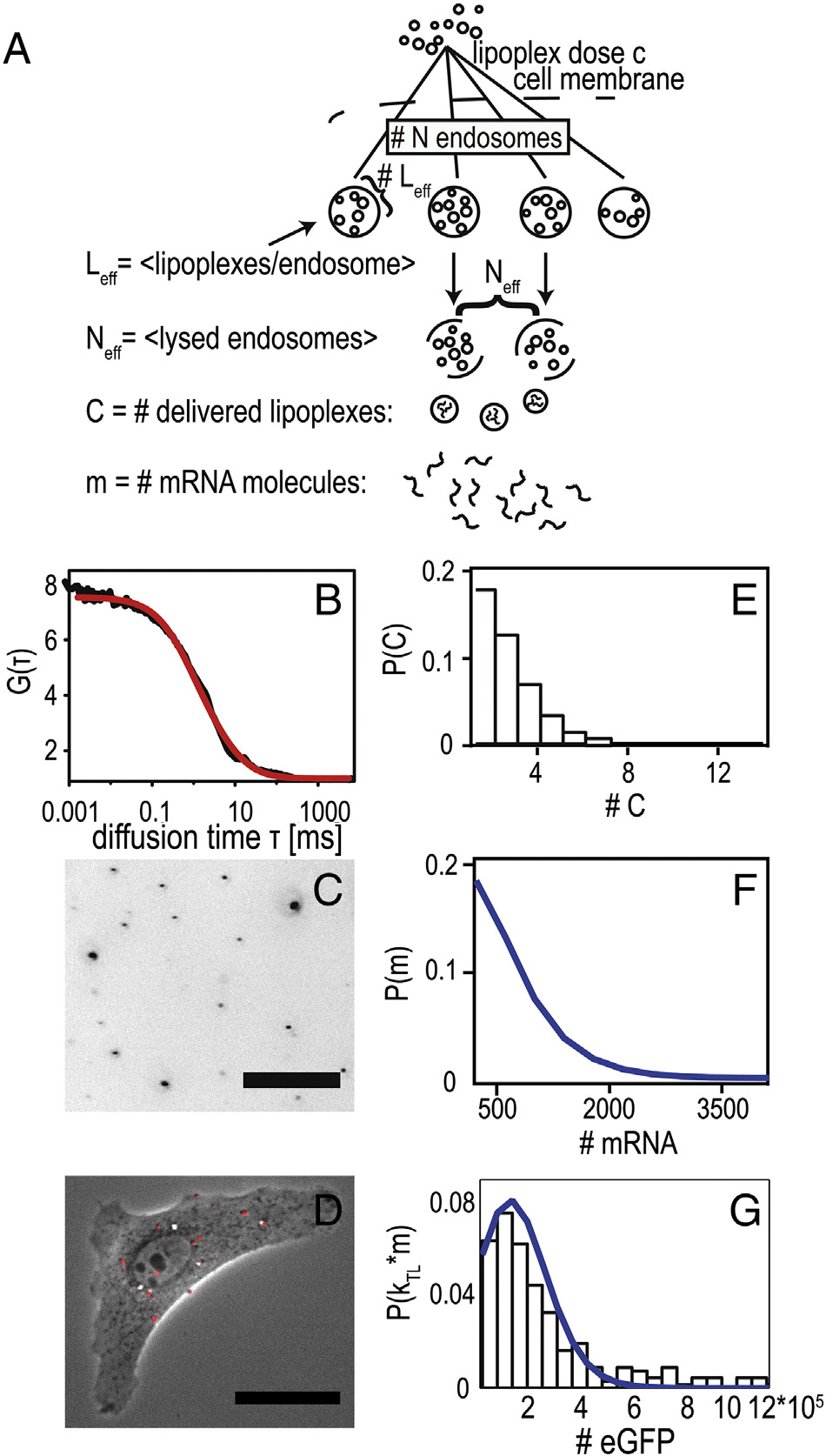
A stochastic delivery model by Numbers
It is generally understood that mRNA as well as pDNA
delivery via artificial, non-viral vectors is stochastic anddominated by rare processes. In the case of mRNA transfection,the limiting steps are endosomal uptake, endosomal lysis, andmRNA release from lipoplexes. Here, we ask the questionwhether the measured distribution of expression levels can bereproduced in a stochastic rate model, where each step isassumed to be described by a random process with definedtransition probability. The fact that a large fraction of cells doesnot express eGFP at all indicates that there is a finite probabilitythat no nucleic acid is successfully transferred. Ashows the dose–response curve in terms of the percentage oftransfected cells versus the concentration of mRNA in μg RNAper ml transfection medium. The corresponding distribution ofeGFP expression levels can be seen in B. Data weretaken 25 h after transfection using FC. The number oftransfected cells monotonically increases with mRNA dosage.
It is instructive to describe the transfection process in terms ofnumber of lipoplexes: Lipoplexes form when cationic lipidliposomes are complexed with nucleic acid. Each lipoplexcontains a large average number of mRNA molecules (asdiscussed below). Hence, the delivery of a single lipoplex resultsin a burst of eGFP expression. If lipoplexes were delivered by
Figure 6. Two-step stochastic mRNA delivery model. (
overcoming a single barrier, the dose–response function would
A) Schematic drawing
of the stochastic uptake of lipoplexes by endosomes, lysis of the endosomes,
be described by a Poisson-like process as represented by the
and release of the mRNA load by lipoplexes. The model reproduces the
dashed line in , A (see Supplementary). In this case, the
dose–response relation shown in Figure 6, A. (B) Fluorescence autocorre-
average number of effectively delivered lipoplexes would be
lation function of lipoplexes showing an average hydrodynamic radius of
〈C〉SP = 0.5. However, as shown in , A, the fraction of
Rhydr. = 60 nm. (C) Fluorescence image of fluorescently labeled mRNA
transfected cells can be more closely described by a chain of two
lipoplexes adsorbed to a petri dish at the concentration that was used for time-
successive Poisson processes. In this case, the response does not
lapse transfection experiments (dose: 1 μg/ml mRNA). Image analysis led toa typical lipoplex density of order 4000/mm2 corresponding to about 4–
rise up to 100% at large mRNA concentration, which is due to
8 lipoplexes per cell (intensity scale inverted for clarity, scale bar 25 μm). (D)
the fact that the two Poisson processes are sequential. A physical
Typical A549 cell five hours after transfection with fluorescently labeled
interpretation of such a chain of events is shown in A: The
mRNA-lipoplexes (shown in red, scale bar 25 μm). (E) Predicted distribution
scheme shows endosomal uptake of lipoplexes, endosomal lysis,
of delivered lipoplexes derived from the dose–response relation. (F)
and mRNA release from lipoplexes. It is assumed that N
Predicted distribution of delivered mRNA molecules, based on an average
endosomes are stochastically loaded with a small number of lipo-
of 350 mRNA molecules per lipoplex. (G) Experimental probability
distribution of expression rates (kTL · m0, black bars) derived from single-
eff, and that subsequently a small fraction of endosomes,
cell data. Blue line indicates best fit of mRNA distribution to the expression
Neff, undergoes lysis. These two stochastic steps are modeled as
distribution, yielding an approximate translation rate of kTL = 170/h.
Poisson processes and determine the number of deliveredlipoplexes, C. If we assume the lipoplex load Leff to be propor-tional to the mRNA concentration, i.e. L
eff = λ⋅cmRNA, we obtain a
that an average of 〈C〉 = Neff Leff = 2 successfully delivered
two-parameter expression for the dose–response function (see
complexes is obtained. To demonstrate that such a surprisingly
, A and Supplementary). The best fit yields Neff = 0.9 and
small number of effectively delivered lipoplexes is realistic, we
λ = 1.1 μg-1, meaning that at the highest dose of 2 μg, an effective
assessed the average number of lipoplexes resting on a single cell in
number of Leff = 2.2 lipoplexes are contained per endosome and
an experiment. At a dose of 1 μg mRNA and after one hour
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
incubation time, we found a lipoplex surface density of about
can predict the transient course of therapeutic efficacy of mRNA
4000/mm2, corresponding to an average of 4–8 lipoplexes per
therapeutics in preclinical studies. For example, the development
cell (C). This number is strongly dependent on
of improved capping sequences of mRNA vectors can be carried
incubation time due to the diffusion limited transport of the
out using destabilized eGFP variants. In this case, the protein
lipoplexes. After five hours of incubation, the number of
level decreases substantially faster and long observation times
lipoplexes doubles as seen in , D. We can safely assume
causing experimental difficulties can be circumvented (see
that almost all lipoplexes that hit the cell surface will be taken
Figure S7, Supplementary). Based on kinetic rates obtained in
up by endocytosis over time as reported by How-
such studies, the time-course of arbitrary gene products with
ever, not every endosome releases its lipoplex cargo into the
longer half-life times can be inferred. In this context, it should be
cytosol. We find that a lysis rate of about 25–50% leads to
noted that the half-life of about 12 hours for eGFP determined
accordance of the experimental dose–response relation with the
from single-cell tracks is shorter than previously reported in
above theoretical estimate.
ensemble measurements, which necessarily average over the
A single lipoplex contains an average of 〈m〉 = 350 mRNA
somewhat heterogeneous timing of whole We
molecules. This number is derived knowing the size and packing
also showed that the cell-to-cell variability in the expression
density of lipoplexes (see Supplementary). The mRNA lipo-
levels is well described by a two-step Poisson process. The two-
plexes used here exhibit an average hydrodynamic radius of
step stochastic model is capable of reproducing the measured
60 nm as measured by fluorescence correlation spectroscopy
dose–response curve consistently with the statistical distribution
(FCS) , B). The structure and packing density have
of expression rates. However, it is limited to transfection in vitro
been measured previously using small angle X-ray scattering
and provides only an approximate description of the underlying
, E shows the theoretical distribution of
delivery cascade. The most important element provided by our
delivered lipoplexes based on the double-Poisson model and the
model is the account of quantal delivery of mRNA in form of
mRNA dose that was used for these experiments (1 μg). If this
lipoplexes, which is in quantitative agreement with the measured
distribution is multiplied with the number of mRNA molecules
distribution functions. The small number of successfully
per lipoplex, we obtain the theoretical distribution of mRNA per
delivered lipoplexes per cell is the key to understanding the
cell as shown in , F. It is noteworthy that the theoretical
stochastic outcome of transfection experiments that inherently
distribution (, G, blue curve) is in very satisfying
allow a finite number of non-transfected cells. More refined
agreement with the shape of the experimental distribution
modeling has to be done to picture the dynamics of transfection
(G, black bars) of expression rates. Comparing the
and to reproduce the onset time distribution. Here, computational
theoretical mRNA distribution with the actually measured
representation of size-dependent uptake rates, the nature of
distribution of expression rates, kTL · m0, we find kTL = 170/h.
endosome lysis, and intracellular diffusion need to be solved.
This translation rate, which emerges from the analysis of single-
Furthermore, computational modeling of extracellular delivery,
cell expression rates, is in the range of independently published
mimicking in vivo situations, needs to be advanced to gain
values of translation
impact on translational medicine.
In our experiments, the single-cell time-courses of mRNA-
mediated transfection showed excellent agreement with the
standard biochemical rate model of translation. Hence, single-cell analysis enables direct determination of expression rates as
We studied the expression kinetics of eGFP following
well as decay rates for both mRNA and eGFP with great
transfection mediated by mRNA and pDNA. While pDNA
accuracy and provides a quantitative foundation for kinetic
complexes have to enter the nucleus, mRNA molecules released
studies on mRNA translational regulation as for example RNA
from mRNA lipoplexes can be translated immediately after
interference. The fact that mRNA transfection exhibits a narrow
endosomal escape. Consequently, mRNA-induced expression is
time window of delivery is beneficial for kinetic studies. This
profoundly earlier and more homogeneously timed than pDNA-
advantage should be of practical importance for future time-
induced expression. This behavior is generic and similar onset
resolved studies on siRNA knockdown and RNA constructs for
time distributions are observed e.g. for HeLa and MDCKII cells
programmed gene regulatory operations.
(data not shown). The high transfection efficiencies for pDNAtransfected cells as compared to mRNA transfected cells might
be a result of size-dependent lipoplex uptake that has beenreported We determined the pDNA-lipoplexes to
We thank Carsten Rudolph for the friendly gift of the vector
be about 230 nm in diameter (data not shown), as opposed to
pSTI-A120, Svenja Lippok for FCS measurements, David Smith
120 nm for mRNA-lipoplexes. The narrow timing of mRNA
for proof-reading of the manuscript, and Maria P. Dobay for
expression onset at approximately 3 hours post-transfection is in
agreement with the observed timing found for endosomal uptakeand release in single-particle tracking Therefore, themRNA expression onset distribution might serve as a valuable
Appendix A. Supplementary data
indicator for the endosomal release time distribution and couldbe useful for the advancement of artificial endosomolytic agents.
Supplementary data to this article can be found online at
Furthermore, our data imply that mRNA expression modeling
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
20. Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R,
Benenson Y. A universal RNAi-based logic evaluator that operates in
1. Tavernier G, Andries O, Demeester J, Sanders NN, De Smedt SC,
mammalian cells. Nat Biotechnol 2007;25(7):795-801.
Rejman J. mRNA as gene therapeutic: how to control protein expression.
21. Win MN, Smolke CD. Higher-order cellular information processing with
J Control Release 2011;150(3):238-47.
synthetic RNA devices. Science 2008;322(5900):456-60.
2. Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Current
22. Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally
prospects for mRNA gene delivery. Eur J Pharm Biopharm 2009;
designed logic integration of regulatory signals in mammalian cells. Nat
3. Andries O, De Filette M, Rejman J, De Smedt SC, Demeester J, Van
23. Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input
Poucke M, et al. Comparison of the gene transfer efficiency of mRNA/
RNAi-based logic circuit for identification of specific cancer cells.
GL67 and pDNA/GL67 complexes in respiratory cells. Mol Pharm
24. Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven
4. Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE.
engineering of RNA devices to quantitatively program gene expression.
Synthesis and properties of mRNAs containing the novel "anti-reverse"
cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)
25. Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA
GpppG. RNA (New York, NY) 2001;7(10):1486-95.
transfection. Proc Natl Acad Sci U S A 1989;86(16):6077-81.
5. Jemielity J, Stepinski J, Jaremko M, Haber D, Stolarski R, Rhoads RE,
26. Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-
et al. Synthesis of novel mRNA 5¢ cap-analogues: dinucleoside P1,
mediated RNA delivery: a novel approach for enhanced transfection of
P3-Tri-, P1, P4-Tetra-, and P1, P5-Pentaphosphates. Nucleosides
primary and post-mitotic cells. Nucleic Acids Res 2001;29(18):3882-91.
Nucleotides Nucleic Acids 2003;22(5–8):691-4.
27. Rejman J, Tavernier G, Bavarsad N, Demeester J, De Smedt SC. mRNA
6. Zohra FT, Chowdhury EH, Tada S, Hoshiba T, Akaike T. Effective
transfection of cervical carcinoma and mesenchymal stem cells mediated
delivery with enhanced translational activity synergistically accelerates
by cationic carriers. J Control Release 2010;147(3):385-91.
mRNA-based transfection. Biochem Biophys Res Commun 2007;358(1):
28. Debus H, Baumhof P, Probst J, Kissel T. Delivery of messenger RNA
using poly(ethylene imine)-poly(ethylene glycol)-copolymer blends for
7. Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, et al.
polyplex formation: biophysical characterization and in vitro transfection
Phosphorothioate cap analogs increase stability and translational
properties. J Control Release 2010;148(3):334-43.
efficiency of RNA vaccines in immature dendritic cells and induce
29. Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR.
superior immune responses in vivo. Gene Ther 2010;17(8):961-71.
Three-dimensional imaging of lipid gene-carriers: membrane charge
8. Pesole G, Grillo G, Larizza A, Liuni S. The untranslated regions of
density controls universal transfection behavior in lamellar cationic
eukaryotic mRNAs: structure, function, evolution and bioinformatic
liposome-DNA complexes. Biophys J 2003;84(5):3307-16.
tools for their analysis. Brief Bioinform 2000;1(3):236-49.
30. Chan C-L, Majzoub RN, Shirazi RS, Ewert KK, Chen Y-J, Liang KS, et
9. Kormann MSD, Hasenpusch G, Aneja MK, Nica G, Flemmer AW,
al. Endosomal escape and transfection efficiency of PEGylated cationic
Herber-Jonat S, et al. Expression of therapeutic proteins after delivery of
liposome–DNA complexes prepared with an acid-labile PEG-lipid.
chemically modified mRNA in mice. Nat Biotech 2011;29(2):154-7.
10. Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, et al.
31. Safinya CR, Ewert K, Ahmad A, Evans HM, Raviv U, Needleman DJ, et al.
Modification of antigen-encoding RNA increases stability, translational
Cationic liposome–DNA complexes: from liquid crystal science to gene
efficacy, and T-cell stimulatory capacity of dendritic cells. Blood
delivery applications. Philos Transact R Soc A, Math Phys Eng Sci
11. Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular
32. Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic
and molecular barriers to gene transfer by a cationic lipid. J Biol Chem
gene delivery vector design properties. Mol Ther 2001;4(5):438-46.
33. Dinh AT, Pangarkar C, Theofanous T, Mitragotri S. Understanding
12. Wilke M, Fortunati E, vandenBroek M, Hoogeveen AT, Scholte BJ.
intracellular transport processes pertinent to synthetic gene delivery via
Efficacy of a peptide-based gene delivery system depends on mitotic
stochastic simulations and sensitivity analyses. Biophys J 2007;92(3):831-46.
activity. Gene Ther 1996;3(12):1133-42.
34. Kamiya H, Akita H, Harashima H. Pharmacokinetic and pharmacody-
13. Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E. Cell cycle
namic considerations in gene therapy. Drug Discov Today 2003;8(21):
dependence of gene transfer by lipoplex polyplex and recombinant
adenovirus. Gene Ther 2000;7(5):401-7.
35. Zou S, Scarfo K, Nantz MH, Hecker JG. Lipid-mediated delivery of
14. Mockey M, Goncalves C, Dupuy FP, Lemoine FM, Pichon C, Midoux P.
RNA is more efficient than delivery of DNA in non-dividing cells. Int J
mRNA transfection of dendritic cells: synergistic effect of ARCA
mRNA capping with Poly(A) chains in cis and in trans for a high protein
36. Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol
expression level. Biochem Biophys Res Commun 2006;340(4):1062-8.
15. Kuhn AN, Diken M, Kreiter S, Vallazza B, Türeci Ö, Sahin U.
37. Schwake G, Youssef S, Kuhr JT, Gude S, David MP, Mendoza E, et al.
Determinants of intracellular RNA pharmacokinetics: implications for
Predictive modeling of non-viral gene transfer. Biotechnol Bioeng
RNA-based immunotherapeutics. RNA Biol 2011;8(1):35-43.
16. De Haes W, Van Mol G, Merlin C, De Smedt SC, Vanham G, Rejman J.
38. Summers HD, Rees P, Holton MD, Rowan Brown M, Chappell SC,
Internalization of mRNA lipoplexes by dendritic cells. Mol Pharm
Smith PJ, et al. Statistical analysis of nanoparticle dosing in a dynamic
cellular system. Nat Nano 2011;6(3):170-4.
17. Arthur JF, Butterfield LH, Roth MD, Bui LA, Kiertscher SM, Lau R,
39. Grudzien E, Stepinski J, Jankowska-Anyszka M, Stolarski R, Darzyn-
et al. A comparison of gene transfer methods in human dendritic cells.
kiewicz E, Rhoads RE. Novel cap analogs for in vitro synthesis of
Cancer Gene Ther 1997;4(1):17-25.
mRNAs with high translational efficiency. RNA (New York, NY.
18. Strobel I, Berchtold S, Gotze A, Schulze U, Schuler G, Steinkasserer A.
Human dendritic cells transfected with either RNA or DNA encoding
40. Youssef S, Gude S, Rädler JO. Automated tracking in live-cell time-
influenza matrix protein M1 differ in their ability to stimulate cytotoxic T
lapse movies. Integr Biol 2011;3(11):1095-101.
lymphocytes. Gene Ther 2000;7(23):2028-35.
41. de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris
19. Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic
M, et al. Cellular dynamics of EGF receptor-targeted synthetic viruses.
acid logic circuits. Science 2006;314(5805):1585-8.
Mol Ther 2007;15(7):1297-305.
C. Leonhardt et al / Nanomedicine: Nanotechnology, Biology, and Medicine xx (2014) xxx–xxx
42. Sacchetti A, El Sewedy T, Nasr AF, Alberti S. Efficient GFP mutations
45. Rädler JO, Koltover I, Salditt T, Safinya CR. Structure of DNA-cationic
profoundly affect mRNA transcription and translation rates. FEBS Lett
liposome complexes: DNA intercalation in multilamellar membranes in
distinct interhelical packing regimes. Science 1997;275(5301):810-4.
43. Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene
46. Rädler JO, Koltover I, Jamieson A, Salditt T, Safinya CR. Structure and
expression. Nature 2003;422(6932):633-7.
interfacial aspects of self-assembled cationic lipid−DNA gene carrier
44. Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, et al. Generation of
complexes§. Langmuir 1998;14(15):4272-83.
destabilized green fluorescent protein as a transcription reporter. J Biol
47. Ross PC, Hui SW. Lipoplex size is a major determinant of in vitro
lipofection efficiency. Gene Ther 1999;6(4):651-9.
Source: http://www.thomassligon.info/Documents/PublicationFiles/Leonhardt-2013.pdf
Rapid detection of genetically modified organisms on a continuous-flow polymerase chain reaction microfluidics
Analytical Biochemistry 385 (2009) 42–49 Contents lists available at Analytical Biochemistry Rapid detection of genetically modified organisms on a continuous-flowpolymerase chain reaction microfluidics Yuyuan Li, Da Xing *, Chunsun Zhang MOE Key Laboratory of Laser Life Science and Institute of Laser Life Science, South China Normal University, No. 55, Zhongshan Avenue West, Tianhe District,Guangzhou 510631, People's Republic of China
tpoly.edu.gh
TAKORADI POLYTECHNIC JOURNAL OF TECHNOLOGY AN OFFICIAL JOURNAL OF TAKORADI POLYTECHNIC Volume 3, Number 1, April 2014 Published by TAKORADI POLYTECHNIC TAKORADI, GHANA, WEST AFRICA © Takoradi Polytechnic Journal of Technology All rights reserved; no part of this publication may be reproduced, stored, in a retrieval system, transmitted